Calibrating GPs using ABC¶
[1]:
import os
# Ignore my broken HDF5 install...
os.putenv("HDF5_DISABLE_VERSION_CHECK", '1')
[2]:
import pandas as pd
import cis
import iris
from utils import get_aeronet_data, get_bc_ppe_data
from esem.utils import validation_plot, plot_parameter_space, get_random_params, ensemble_collocate
from esem import gp_model
from esem.abc_sampler import ABCSampler, constrain
import iris.quickplot as qplt
import matplotlib.pyplot as plt
%matplotlib inline
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\h5py\__init__.py:40: UserWarning: h5py is running against HDF5 1.10.6 when it was built against 1.10.5, this may cause problems
'{0}.{1}.{2}'.format(*version.hdf5_built_version_tuple)
Read in the parameters and observables¶
[3]:
aaod = get_aeronet_data()
print(aaod)
Ungridded data: Absorption_AOD440nm / (1)
Shape = (10098,)
Total number of points = 10098
Number of non-masked points = 10098
Long name = Absorption_AOD440nm
Standard name = None
Units = 1
Missing value = -999.0
Range = (5.1e-05, 0.47236)
History =
Coordinates:
longitude
Long name =
Standard name = longitude
Units = degrees_east
Missing value = None
Range = (-155.576755, 141.3407)
History =
latitude
Long name =
Standard name = latitude
Units = degrees_north
Missing value = None
Range = (-35.495807, 79.990278)
History =
time
Long name =
Standard name = time
Units = days since 1600-01-01 00:00:00
Missing value = None
Range = (cftime.DatetimeGregorian(2017, 1, 1, 12, 0, 0, 0), cftime.DatetimeGregorian(2018, 1, 2, 12, 0, 0, 0))
History =
[4]:
# Read in the PPE parameters, AAOD and DRE
ppe_params, ppe_aaod, ppe_dre = get_bc_ppe_data(dre=True)
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\iris\__init__.py:249: IrisDeprecation: setting the 'Future' property 'netcdf_promote' is deprecated and will be removed in a future release. Please remove code that sets this property.
warn_deprecated(msg.format(name))
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\iris\__init__.py:249: IrisDeprecation: setting the 'Future' property 'netcdf_promote' is deprecated and will be removed in a future release. Please remove code that sets this property.
warn_deprecated(msg.format(name))
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\iris\__init__.py:249: IrisDeprecation: setting the 'Future' property 'netcdf_promote' is deprecated and will be removed in a future release. Please remove code that sets this property.
warn_deprecated(msg.format(name))
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\iris\__init__.py:249: IrisDeprecation: setting the 'Future' property 'netcdf_promote' is deprecated and will be removed in a future release. Please remove code that sets this property.
warn_deprecated(msg.format(name))
[5]:
# Take the annual mean of the DRE
ppe_dre, = ppe_dre.collapsed('time', iris.analysis.MEAN)
WARNING:root:Creating guessed bounds as none exist in file
WARNING:root:Creating guessed bounds as none exist in file
WARNING:root:Creating guessed bounds as none exist in file
WARNING:root:Creating guessed bounds as none exist in file
Collocate the model on to the observations¶
[6]:
col_ppe_aaod = ensemble_collocate(ppe_aaod, aaod)
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\ma\core.py:3225: FutureWarning: Using a non-tuple sequence for multidimensional indexing is deprecated; use `arr[tuple(seq)]` instead of `arr[seq]`. In the future this will be interpreted as an array index, `arr[np.array(seq)]`, which will result either in an error or a different result.
dout = self.data[indx]
[7]:
n_test = 8
X_test, X_train = ppe_params[:n_test], ppe_params[n_test:]
Y_test, Y_train = col_ppe_aaod[:n_test], col_ppe_aaod[n_test:]
[8]:
Y_train
[8]:
| Absorption Optical Thickness - Total 550Nm (1) | job | obs |
|---|---|---|
| Shape | 31 | 10098 |
| Dimension coordinates | ||
| job | x | - |
| obs | - | x |
| Auxiliary coordinates | ||
| latitude | - | x |
| longitude | - | x |
| time | - | x |
Setup and run the models¶
Explore different model choices¶
[9]:
from esem.utils import leave_one_out, prediction_within_ci
from scipy import stats
import numpy as np
from esem.data_processors import Log
res_l = leave_one_out(X_train, Y_train, model='GaussianProcess', data_processors=[Log(constant=0.1)], kernel=['Linear', 'Exponential', 'Bias'])
r2_values_l = [stats.linregress(x.data.compressed(), y.data[:, ~x.data.mask].flatten())[2]**2 for x,y,_ in res_l]
ci95_values_l = [prediction_within_ci(x.data.flatten(), y.data.flatten(), v.data.flatten())[2].sum()/x.data.count() for x,y,v in res_l]
print("Mean R^2: {:.2f}".format(np.asarray(r2_values_l).mean()))
print("Mean proportion within 95% CI: {:.2f}".format(np.asarray(ci95_values_l).mean()))
res = leave_one_out(X_train, Y_train, model='GaussianProcess', kernel=['Linear', 'Bias'])
r2_values = [stats.linregress(x.data.flatten(), y.data.flatten())[2]**2 for x,y,v in res]
ci95_values = [prediction_within_ci(x.data.flatten(), y.data.flatten(), v.data.flatten())[2].sum()/x.data.count() for x,y,v in res]
print("Mean R^2: {:.2f}".format(np.asarray(r2_values).mean()))
print("Mean proportion within 95% CI: {:.2f}".format(np.asarray(ci95_values).mean()))
# Note that while the Log pre-processing leads to slightly better R^2, the model is under-confident and
# has too large uncertainties which would adversley effec our implausibility metric.
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
c:\users\duncan\pycharmprojects\gcem\esem\data_processors.py:67: RuntimeWarning: invalid value encountered in log
return np.log(data + self.constant)
Mean R^2: 1.00
Mean proportion within 95% CI: 1.00
Mean R^2: 0.99
Mean proportion within 95% CI: 0.94
Build final model¶
[10]:
model = gp_model(X_train, Y_train, kernel=['Linear', 'Bias'])
[11]:
model.train()
[12]:
m, v = model.predict(X_test.values)
[13]:
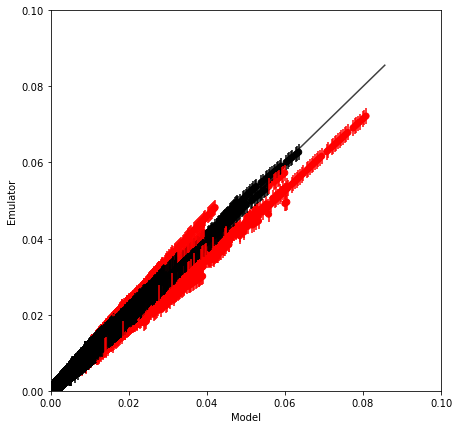
validation_plot(Y_test.data.flatten(), m.data.flatten(), v.data.flatten(),
minx=0, maxx=0.1, miny=0., maxy=0.1)
Proportion of 'Bad' estimates : 5.52%
C:\Users\duncan\miniconda3\envs\gcem_dev\lib\site-packages\numpy\core\_asarray.py:83: UserWarning: Warning: converting a masked element to nan.
return array(a, dtype, copy=False, order=order)
Sample and constrain the models¶
Emulating 1e6 sample points directly would require 673 Gb of memory so we can either run 1e6 samples for each point, or run the constraint everywhere, but in batches. Here we do the latter, optioanlly on the GPU, using the ‘naive’ algorithm for calculating the running mean and variance of the various properties.
The rejection sampling happens in a similar manner so that only as much memory as is used for one batch is ever used.
[14]:
# In this case
sample_points = pd.DataFrame(data=get_random_params(3, int(1e6)), columns=X_train.columns)
[15]:
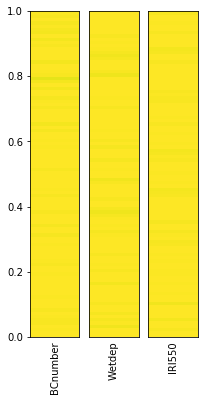
# Note that smoothing the parameter distribution can be slow for large numbers of points
plot_parameter_space(sample_points, fig_size=(3,6), smooth=False)
[16]:
# Setup the sampler to compare against our AeroNet data
sampler = ABCSampler(model, aaod, obs_uncertainty=0.5, repres_uncertainty=0.5)
[17]:
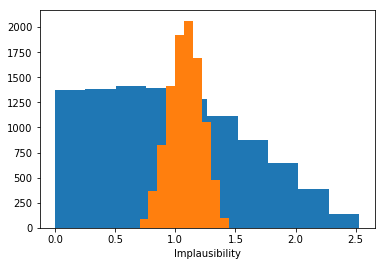
# Calculate the implausibilty for each sample against each observation - note this can be very large so we only sample a fraction!
implaus = sampler.get_implausibility(sample_points[::100], batch_size=1000)
# The implausibility distributions for different observations can be very different.
_ = plt.hist(implaus.data[:, 1400])
_ = plt.hist(implaus.data[:, 14])
plt.gca().set(xlabel='Implausibility')
100%|##########| 10000/10000 [00:05<00:00, 1859.15sample/s]
[17]:
[Text(0.5,0,'Implausibility')]
[18]:
# Find the valid samples in our full 1million samples by comparing against a given tolerance and threshold
valid_samples = sampler.batch_constrain(sample_points, batch_size=1000, tolerance=.1)
print("Remaining points: {}".format(valid_samples.sum()))
100%|##########| 1000000/1000000 [05:23<00:00, 2972.65sample/s]Remaining points: 729607
[19]:
# Plot the reduced parameter distribution
constrained_sample = sample_points[valid_samples]
plot_parameter_space(constrained_sample, fig_size=(3,6))

[20]:
# We can also easily plot the joint distributions
# Only plot every one in 100 points as scatter plots with large numbers of points are slow...
import matplotlib
# Mimic Seaborn scaling without requiring the whole package
scale = 1.5
matplotlib.rcParams['font.size'] = 12 * scale
matplotlib.rcParams['axes.labelsize'] = 12 * scale
matplotlib.rcParams['axes.titlesize'] = 12 * scale
matplotlib.rcParams['xtick.labelsize'] = 11 * scale
matplotlib.rcParams['ytick.labelsize'] = 11 * scale
matplotlib.rcParams['lines.linewidth'] = 1.5 * scale
matplotlib.rcParams['lines.markersize'] = 6 * scale
#
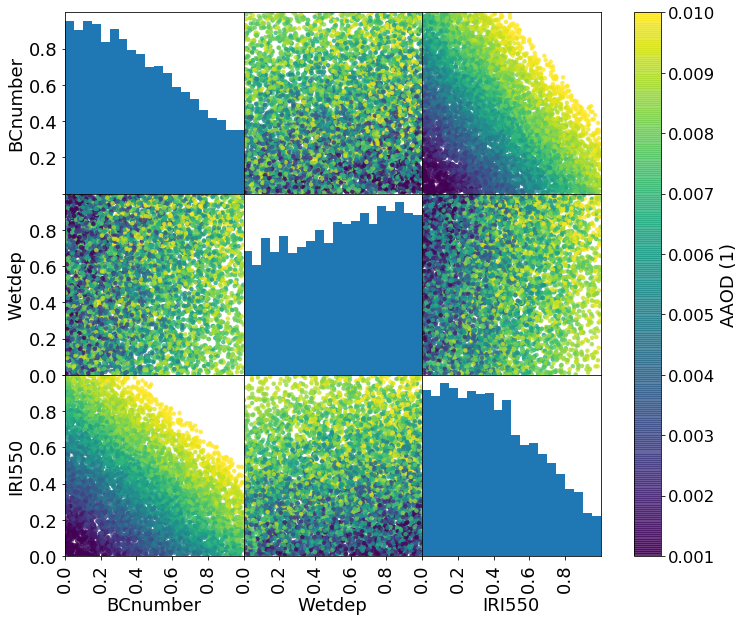
m, _ = model.predict(constrained_sample[::100].values)
Zs = m.data
# Plot the emulated AAOD value (averaged over observation locations) for each point
grr = pd.plotting.scatter_matrix(constrained_sample[::100], c=Zs.mean(axis=1), figsize=(12, 10), marker='o',
hist_kwds={'bins': 20,}, s=20, alpha=.8, vmin=1e-3, vmax=1e-2, range_padding=0.,
density_kwds={'range': [[0., 1.], [0., 1.]], 'colormap':'viridis'},
)
# Matplotlib dragons...
grr[0][0].set_yticklabels([0.2, 0.4, 0.6, 0.8], fontsize=12 * scale)
for i in range(2):
grr[i+1][0].set_yticklabels([0.0, 0.2, 0.4, 0.6, 0.8], fontsize=12 * scale)
for i in range(3):
grr[2][i].set_xticks([0.0, 0.2, 0.4, 0.6, 0.8])
grr[2][i].set_xticklabels([0.0, 0.2, 0.4, 0.6, 0.8], fontsize=12 * scale)
plt.colorbar(grr[0][1].collections[0], ax=grr, use_gridspec=True, label='AAOD (1)')
plt.savefig('BCPPE_constrained_params_paper.png', transparent=True)
Explore the uncertainty in Direct Radiative Effect of Aerosol in constrianed sample-space¶
[21]:
dre_test, dre_train = ppe_dre[:n_test], ppe_dre[n_test:]
ari_model = gp_model(X_train, dre_train, name="ARI", kernel=['Linear', 'Bias'])
ari_model.train()
[22]:
# Calculate the mean and std-dev DRE over each set of sample points
unconstrained_mean_ari, unconstrained_sd_ari = ari_model.batch_stats(sample_points, batch_size=1000)
constrained_mean_ari, constrained_sd_ari = ari_model.batch_stats(constrained_sample, batch_size=1000)
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-22-1ce0d8f67b9a> in <module>
1 # Calculate the mean and std-dev DRE over each set of sample points
2
----> 3 unconstrained_mean_ari, unconstrained_sd_ari = ari_model.batch_stats(sample_points, batch_size=1000)
4 constrained_mean_ari, constrained_sd_ari = ari_model.batch_stats(constrained_sample, batch_size=1000)
c:\users\duncan\pycharmprojects\gcem\esem\emulator.py in batch_stats(self, sample_points, batch_size)
112 # TODO: Make sample points optional and just sample from a uniform distribution if not provided
113 mean, sd = _tf_stats(self, tf.constant(sample_points),
--> 114 tf.constant(batch_size, dtype=tf.int64),
115 pbar=tf_tqdm(batch_size=batch_size,
116 total=sample_points.shape[0]))
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\def_function.py in __call__(self, *args, **kwds)
826 tracing_count = self.experimental_get_tracing_count()
827 with trace.Trace(self._name) as tm:
--> 828 result = self._call(*args, **kwds)
829 compiler = "xla" if self._experimental_compile else "nonXla"
830 new_tracing_count = self.experimental_get_tracing_count()
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\def_function.py in _call(self, *args, **kwds)
869 # This is the first call of __call__, so we have to initialize.
870 initializers = []
--> 871 self._initialize(args, kwds, add_initializers_to=initializers)
872 finally:
873 # At this point we know that the initialization is complete (or less
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\def_function.py in _initialize(self, args, kwds, add_initializers_to)
724 self._concrete_stateful_fn = (
725 self._stateful_fn._get_concrete_function_internal_garbage_collected( # pylint: disable=protected-access
--> 726 *args, **kwds))
727
728 def invalid_creator_scope(*unused_args, **unused_kwds):
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\function.py in _get_concrete_function_internal_garbage_collected(self, *args, **kwargs)
2967 args, kwargs = None, None
2968 with self._lock:
-> 2969 graph_function, _ = self._maybe_define_function(args, kwargs)
2970 return graph_function
2971
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\function.py in _maybe_define_function(self, args, kwargs)
3359
3360 self._function_cache.missed.add(call_context_key)
-> 3361 graph_function = self._create_graph_function(args, kwargs)
3362 self._function_cache.primary[cache_key] = graph_function
3363
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\function.py in _create_graph_function(self, args, kwargs, override_flat_arg_shapes)
3204 arg_names=arg_names,
3205 override_flat_arg_shapes=override_flat_arg_shapes,
-> 3206 capture_by_value=self._capture_by_value),
3207 self._function_attributes,
3208 function_spec=self.function_spec,
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\framework\func_graph.py in func_graph_from_py_func(name, python_func, args, kwargs, signature, func_graph, autograph, autograph_options, add_control_dependencies, arg_names, op_return_value, collections, capture_by_value, override_flat_arg_shapes)
988 _, original_func = tf_decorator.unwrap(python_func)
989
--> 990 func_outputs = python_func(*func_args, **func_kwargs)
991
992 # invariant: `func_outputs` contains only Tensors, CompositeTensors,
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\eager\def_function.py in wrapped_fn(*args, **kwds)
632 xla_context.Exit()
633 else:
--> 634 out = weak_wrapped_fn().__wrapped__(*args, **kwds)
635 return out
636
~\miniconda3\envs\gcem_dev\lib\site-packages\tensorflow\python\framework\func_graph.py in wrapper(*args, **kwargs)
975 except Exception as e: # pylint:disable=broad-except
976 if hasattr(e, "ag_error_metadata"):
--> 977 raise e.ag_error_metadata.to_exception(e)
978 else:
979 raise
TypeError: in user code:
TypeError: tf__batch_stats() takes from 2 to 3 positional arguments but 4 were given
[23]:
# The original (unconstrained DRE)
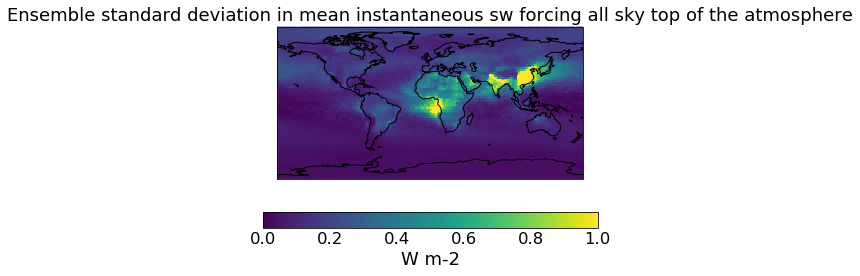
qplt.pcolormesh(unconstrained_sd_ari, vmin=0., vmax=1)
plt.gca().coastlines()
[23]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x2114ad520c8>
[24]:
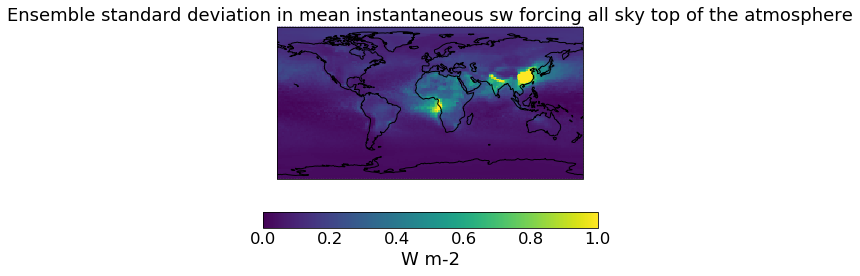
# The constrained DRE
qplt.pcolormesh(constrained_sd_ari, vmin=0., vmax=1)
plt.gca().coastlines()
[24]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x2114a4e35c8>
[25]:
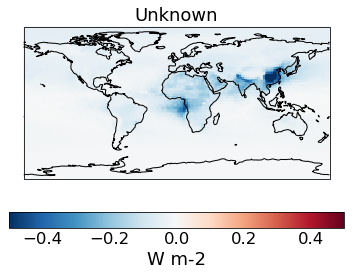
# The change in spread after the constraint is applied
qplt.pcolormesh((constrained_sd_ari-unconstrained_sd_ari), cmap='RdBu_r', vmin=-5e-1, vmax=5e-1)
plt.gca().coastlines()
[25]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x21132beb148>
[ ]: